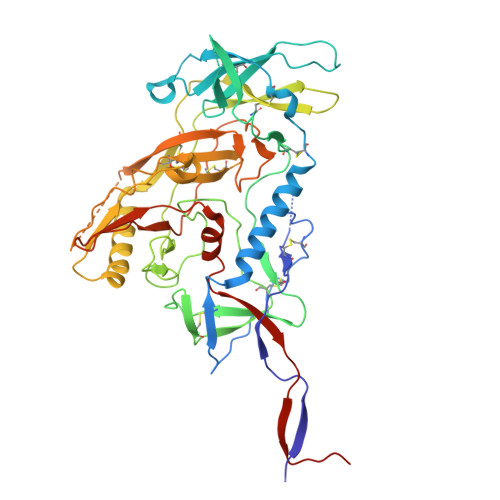
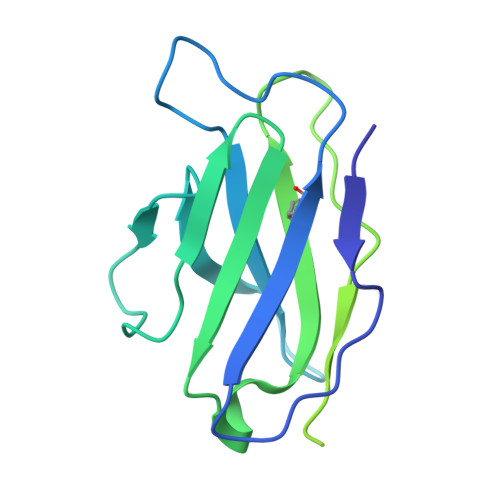
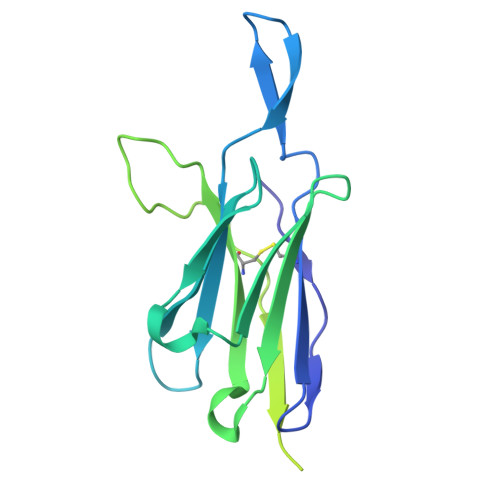
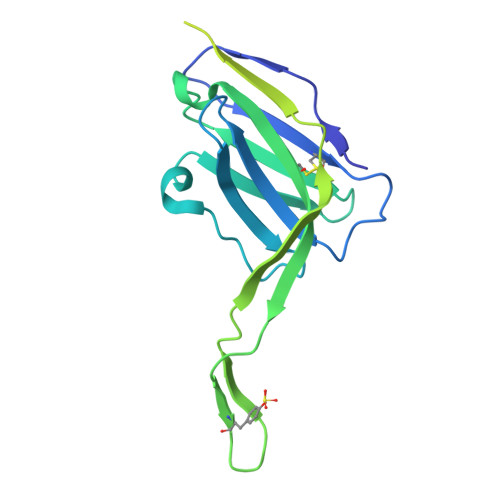
Profiling of HIV-1 elite neutralizer cohort reveals a CD4bs bnAb for HIV-1 prevention and therapy.
Gieselmann, L., DeLaitsch, A.T., Rohde, M., Gruell, H., Kreer, C., Ercanoglu, M.S., Gristick, H.B., Schommers, P., Ahmadov, E., Radford, C., Mazzolini, A., Zhang, L., West Jr., A.P., Worczinski, J., Momot, A., Reichwein, M.L., Knufer, J., Stumpf, R., Mkhize, N.N., Kaldine, H., Bhebhe, S., Deshpande, S., Giovannoni, F., Stefanutti, E., Benigni, F., Havenar-Daughton, C., Corti, D., Kroidl, A., Adhikari, A., Nanfack, A.J., Ambada, G.E., Duerr, R., Maganga, L., William, W., Ntinginya, N.E., Wolf, T., Geldmacher, C., Hoelscher, M., Lehmann, C., Moore, P.L., Mora, T., Walczak, A.M., Gilbert, P.B., Doria-Rose, N.A., Huang, Y., Bloom, J.D., Seaman, M.S., Bjorkman, P.J., Klein, F.(2025) Nat Immunol
- PubMed: 41053396
- DOI: https://doi.org/10.1038/s41590-025-02286-5
- Primary Citation of Related Structures:
8UKI, 8ULR, 8ULS, 8ULT, 8ULU, 9D8V - PubMed Abstract:
Administration of HIV-1 neutralizing antibodies can suppress viremia and prevent infection in vivo. However, clinical use is challenged by envelope diversity and rapid viral escape. Here, we performed single B cell profiling of 32 top HIV-1 elite neutralizers to identify broadly neutralizing antibodies with highest antiviral activity. From 831 expressed monoclonal antibodies, we identified 04_A06, a V H 1-2-encoded broadly neutralizing antibody to the CD4 binding site with remarkable breadth and potency against multiclade pseudovirus panels (geometric mean half-maximal inhibitory concentration = 0.059 µg ml -1 , breadth = 98.5%, 332 strains). Moreover, 04_A06 was not susceptible to classic CD4 binding site escape variants and maintained full viral suppression in HIV-1-infected humanized mice. Structural analyses revealed an unusually long 11-amino-acid heavy chain insertion that facilitates interprotomer contacts with highly conserved residues on the adjacent gp120 protomer. Finally, 04_A06 demonstrated high activity against contemporaneously circulating viruses from the Antibody-Mediated Prevention trials (geometric mean half-maximal inhibitory concentration = 0.082 µg ml -1 , breadth = 98.4%, 191 virus strains), and in silico modeling for 04_A06LS predicted prevention efficacy of >93%. Thus, 04_A06 will provide unique opportunities for effective treatment and prevention of HIV-1 infection.
- Laboratory of Experimental Immunology, Institute of Virology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany.
Organizational Affiliation: