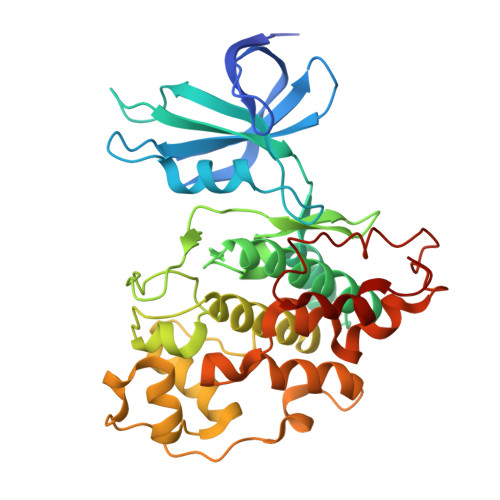
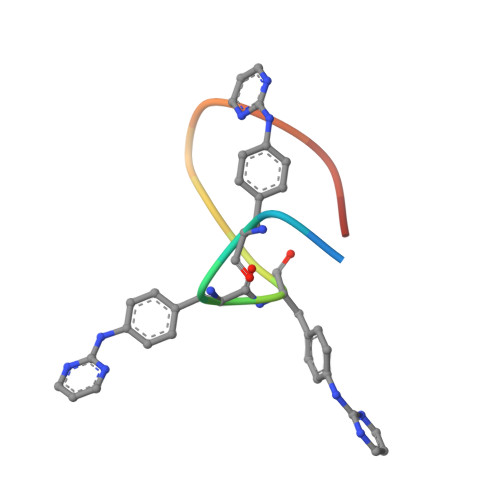
Discovery of Supra-Bivalent GSK3 beta Inhibitory Peptides Containing an ATP-Mimetic Amino Acid.
Haslbock, S., Vinogradov, A.A., Okada, C., Fujimura, N., Aikawa, H., Sengoku, T., Suga, H.(2026) J Am Chem Soc 148: 368-378
- PubMed: 41481598
- DOI: https://doi.org/10.1021/jacs.5c13788
- Primary Citation of Related Structures:
9X2Q, 9X2U, 9X2V, 9X2W, 9X2X, 9X2Y - PubMed Abstract:
De novo discovery of highly selective inhibitors of protein kinases (PKs) remains a significant challenge in drug discovery. Although ATP-site-directed bivalent inhibitors targeting a distinct binding site are an emerging alternative, their rational design is impeded by limited knowledge about allosteric binding sites unique to the target kinase and the complexity of the linker design. Here, we report a strategy that overcomes the above-mentioned issue using a macrocyclic peptide library containing an ATP-mimetic "warhead" amino acid, p -(pyrimidin-2-ylamino)phenylalanine (termed Z), performing the RaPID selection for the discovery of highly potent and selective inhibitors against the kinase GSK3β. The experiment led to the enrichment of macrocycles containing a sequence motif "ZRZ", whose enrichment score log 2 Y was calculated for the identification of high-affinity binders by means of next-generation sequencing analysis. We identified several highly potent inhibitors, with a representative potent peptide (BiS3) showing a K D of 0.3 nM and an IC 50 of 4.8 nM against GSK3β. The X-ray structural analysis revealed the unique binding mode of the peptides with "RZ" mimicking ATP and Mg 2+ in the ATP site. Moreover, the rest of the peptide motifs engaged in multisite interactions via a substrate-competitive site and an unexpected knob-into-hole-like conformation of the macrocycle, and thus, they act as "supra-bivalent" kinase inhibitors. This unique binding mode conferred both potency and selectivity toward GSK3β. This work demonstrated that our strategy using the warhead amino acid Z is effective in obtaining specific GSK3β inhibitors, which is likely expandable to other PK families.
- Department of Chemistry, Graduate School of Science, The University of Tokyo, Tokyo 113-0033, Japan.
Organizational Affiliation: