CryoEM structure of GluK2 bound to glutamate in the shallow desensitized state, composite map
Chang, P.Z., Nami, T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glutamate receptor ionotropic, kainate 2 | 908 | Rattus norvegicus | Mutation(s): 0 Gene Names: Grik2, Glur6 | 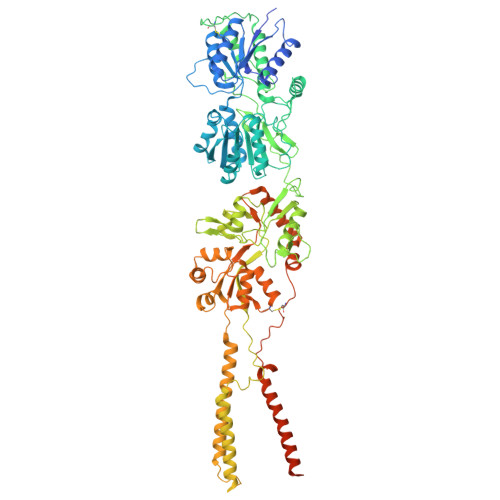 | |
UniProt | |||||
Find proteins for P42260 (Rattus norvegicus) Explore P42260 Go to UniProtKB: P42260 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P42260 | ||||
Glycosylation | |||||
| Glycosylation Sites: 6 | Go to GlyGen: P42260-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AA [auth D] BA [auth D] N [auth A] O [auth A] P [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| GLU Query on GLU | CA [auth D], Q [auth A], T [auth B], X [auth C] | GLUTAMIC ACID C5 H9 N O4 WHUUTDBJXJRKMK-VKHMYHEASA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R35GM147266-01 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 3R35GM147266-01S1 |