Structural insights into human Pol III transcription initiation in action.
Wang, Q., Ren, Y., Jin, Q., Chen, X., Xu, Y.(2025) Nature 643: 1127-1134
- PubMed: 40468065
- DOI: https://doi.org/10.1038/s41586-025-09093-w
- Primary Citation of Related Structures:
9K2G, 9K36, 9K38, 9K39, 9K3B, 9K3U, 9K3V, 9LKT, 9LXN, 9LXO - PubMed Abstract:
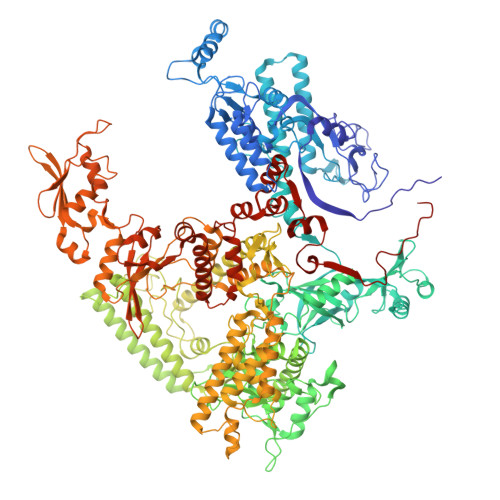
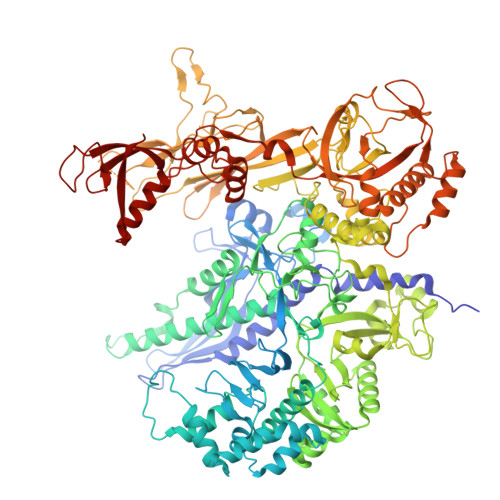
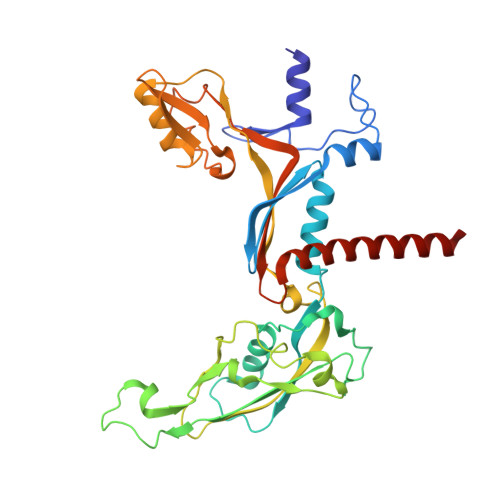
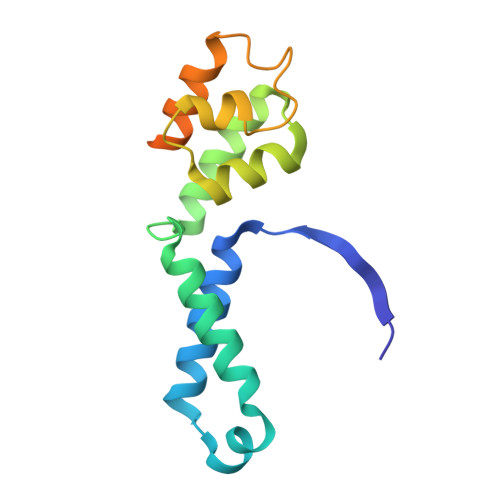
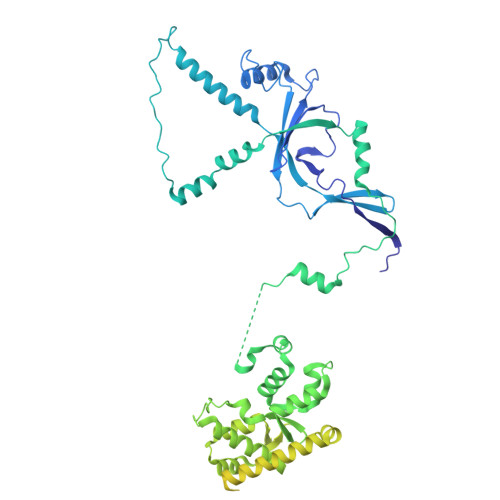
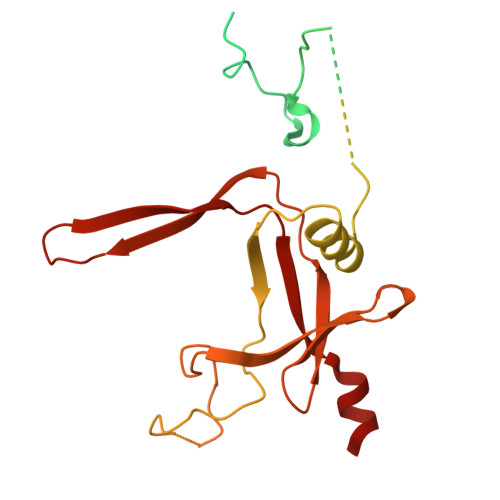
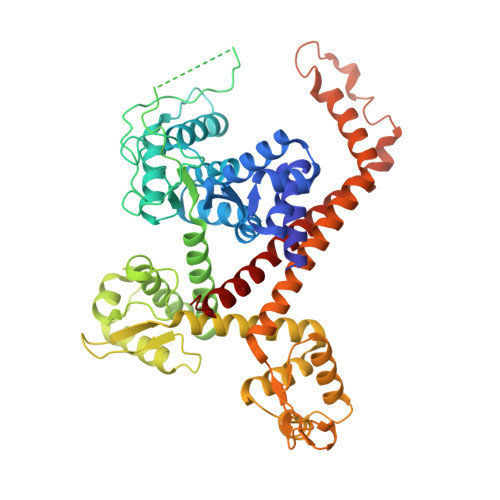
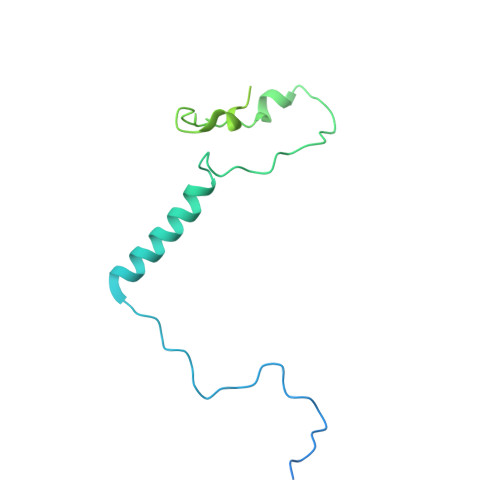
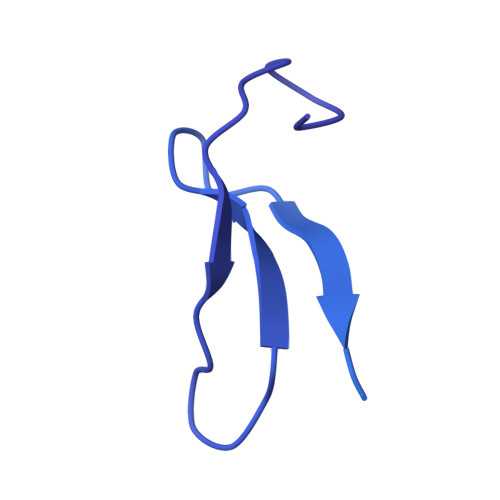
RNA polymerase III (Pol III) transcribes highly demanded RNAs grouped into three types of classical promoters, including type 1 (5S rRNA), type 2 (tRNA) and type 3 (short non-coding RNAs, such as U6, 7SK and RNase H1) promoters 1-7 . While structures of the Pol III preinitiation complex (PIC) 8-11 and elongation complex (EC) 12-16 have been determined, the mechanism underlying the transition from initiation to elongation remains unclear. Here we reconstituted seven human Pol III transcribing complexes (TC4, TC5, TC6, TC8, TC10, TC12 and TC13) halted on U6 promoters with nascent RNAs of 4-13 nucleotides. Cryo-electron microscopy structures captured initially transcribing complexes (ITCs; TC4 and TC5) and ECs (TC6-13). Together with KMnO 4 footprinting, the data reveal extensive modular rearrangements: the transcription bubble expands from PIC to TC5, followed by general transcription factor (GTF) dissociation and abrupt bubble collapse from TC5 to TC6, marking the ITC-EC transition. In TC5, SNAPc and TFIIIB remain bound to the promoter and Pol III, while the RNA-DNA hybrid adopts a tilted conformation with template DNA blocked by BRF2, a TFIIIB subunit. Hybrid forward translocation during ITC-EC transition triggers BRF2-finger retraction, GTF release and transcription-bubble collapse. Pol III then escapes the promoter while GTFs stay bound upstream, potentially enabling reinitiation. These findings reveal molecular insights into Pol III dynamics and reinitiation mechanisms on type 3 promoters of highly demanded small RNAs, with the earliest documented initiation-elongation transition for an RNA polymerase.
- Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, New Cornerstone Science Laboratory, State Key Laboratory of Genetics and Development of Complex Phenotypes, Department of Biochemistry and Biophysics, School of Life Sciences, Shanghai Key Laboratory of Radiation Oncology and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai, China.
Organizational Affiliation: