The structure of a villin-stabilized actin nucleus
Robinson, R.C.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin, alpha skeletal muscle | 377 | Gallus gallus | Mutation(s): 0 Gene Names: ACTA1, ACTA EC: 3.6.4 | 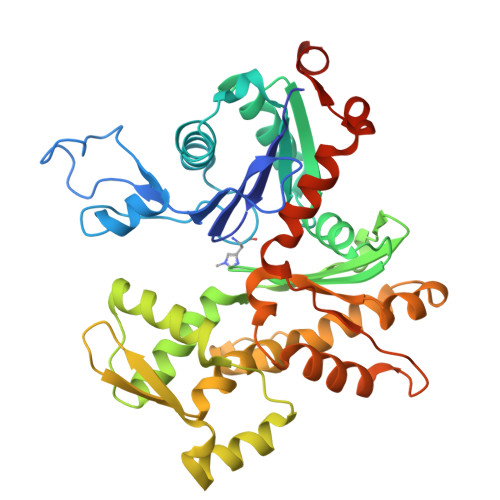 | |
UniProt | |||||
Find proteins for P68139 (Gallus gallus) Explore P68139 Go to UniProtKB: P68139 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68139 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| villin | D [auth v], H [auth V] | 823 | Paralvinella sulfincola | Mutation(s): 0 | 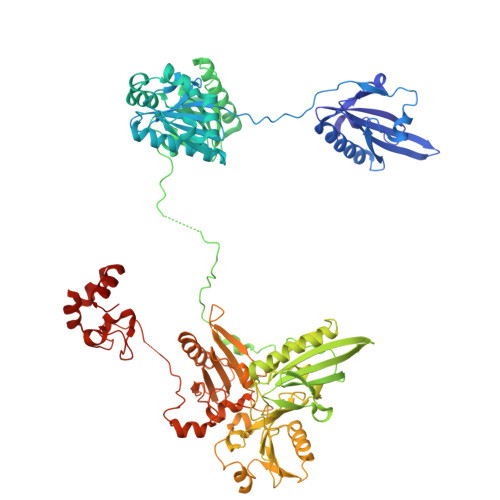 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | BA [auth G] I [auth p] K [auth f] M [auth g] X [auth P] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| SCN Query on SCN | LA [auth V] | THIOCYANATE ION C N S ZMZDMBWJUHKJPS-UHFFFAOYSA-M |  | ||
| CA (Subject of Investigation/LOI) Query on CA | DA [auth V] EA [auth V] FA [auth V] GA [auth V] HA [auth V] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | W [auth v] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG (Subject of Investigation/LOI) Query on MG | AA [auth F] CA [auth G] J [auth p] L [auth f] N [auth g] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| HIC Query on HIC | A [auth p] B [auth f] C [auth g] E [auth P] F A [auth p], B [auth f], C [auth g], E [auth P], F, G | L-PEPTIDE LINKING | C7 H11 N3 O2 |  | HIS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 98.137 | α = 77.95 |
| b = 102.432 | β = 72.59 |
| c = 147.128 | γ = 65.96 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | Moore-Simons GBMF9743 | |
| Human Frontier Science Program (HFSP) | France | RGP0028/2018 |