Structure based Drug design of human Factor VIIa Inhibitors.
Krishnan, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Factor VII light chain | A [auth L] | 142 | Homo sapiens | Mutation(s): 0 Gene Names: F7 EC: 3.4.21.21 | 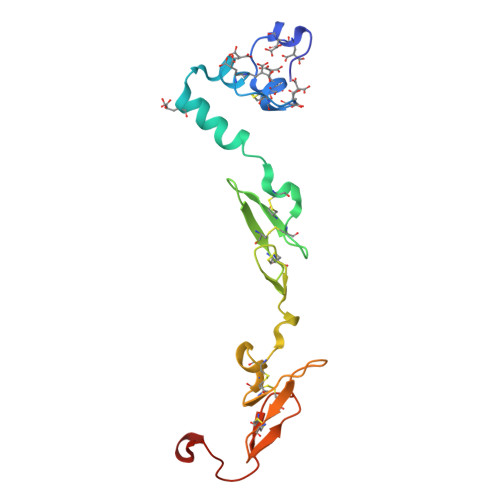 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P08709-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Coagulation factor VII Heavy Chain | B [auth H] | 254 | Homo sapiens | Mutation(s): 0 Gene Names: F7 EC: 3.4.21.21 | 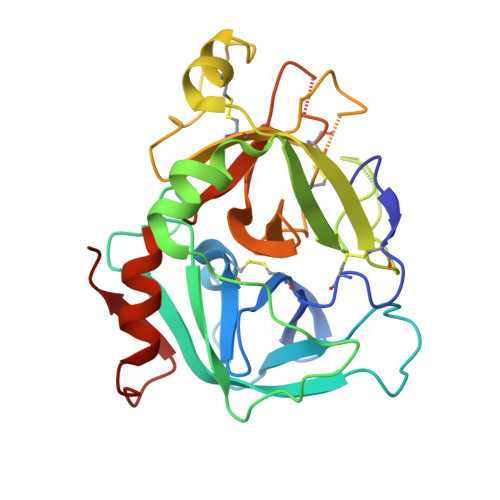 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tissue factor | C [auth T] | 75 | Homo sapiens | Mutation(s): 0 Gene Names: F3 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P13726 (Homo sapiens) Explore P13726 Go to UniProtKB: P13726 | |||||
PHAROS: P13726 GTEx: ENSG00000117525 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13726 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tissue factor | D [auth U] | 116 | Homo sapiens | Mutation(s): 0 Gene Names: F3 | 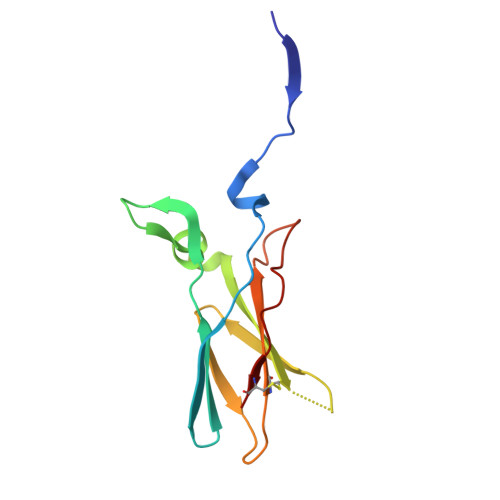 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P13726 (Homo sapiens) Explore P13726 Go to UniProtKB: P13726 | |||||
PHAROS: P13726 GTEx: ENSG00000117525 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13726 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XGX Query on XGX | P [auth H] | (1P)-2'-[(4-carbamimidoylphenyl)carbamoyl]-4'-ethenyl-4-[(2-methylpropyl)carbamoyl][1,1'-biphenyl]-2-carboxylic acid C28 H28 N4 O4 FCTCJAGJUHHZDO-UHFFFAOYSA-N |  | ||
| BGC Query on BGC | E [auth L] | beta-D-glucopyranose C6 H12 O6 WQZGKKKJIJFFOK-VFUOTHLCSA-N |  | ||
| FUC Query on FUC | F [auth L] | alpha-L-fucopyranose C6 H12 O5 SHZGCJCMOBCMKK-SXUWKVJYSA-N |  | ||
| CA Query on CA | G [auth L] H [auth L] I [auth L] J [auth L] K [auth L] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CGU Query on CGU | A [auth L] | L-PEPTIDE LINKING | C6 H9 N O6 |  | GLU |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 70.494 | α = 90 |
| b = 82.87 | β = 90 |
| c = 126.555 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| CrystalClear | data reduction |
| SCALEPACK | data scaling |
| CNX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |