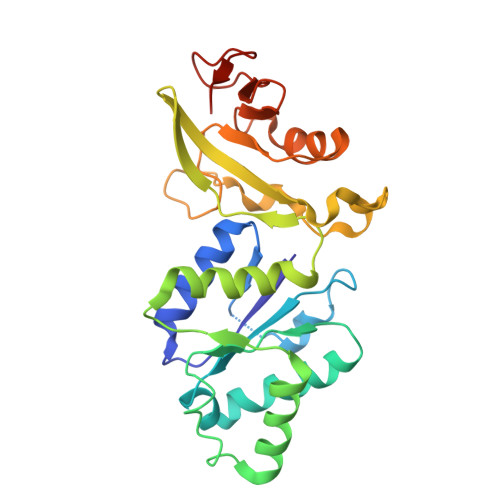
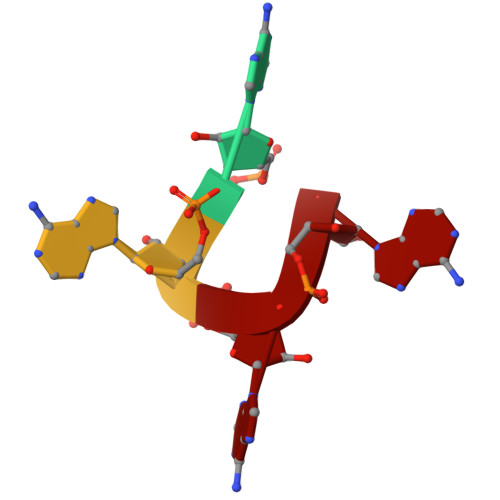
Cat1 forms filament networks to degrade NAD + during the type III CRISPR-Cas antiviral response.
Baca, C.F., Majumder, P., Hickling, J.H., Patel, D.J., Marraffini, L.A.(2025) Science 388: eadv9045-eadv9045
- PubMed: 40208959
- DOI: https://doi.org/10.1126/science.adv9045
- Primary Citation of Related Structures:
9MUD, 9MUE, 9MUO, 9MW9 - PubMed Abstract:
Type III CRISPR-Cas systems defend against viral infection in prokaryotes using an RNA-guided complex that recognizes foreign transcripts and synthesizes cyclic oligo-adenylate (cOA) messengers to activate CARF immune effectors. Here we investigated a protein containing a CARF domain fused Toll/interleukin-1 receptor (TIR) domain, Cat1. We found that Cat1 provides immunity by cleaving and depleting NAD + molecules from the infected host, inducing a growth arrest that prevents viral propagation. Cat1 forms dimers that stack upon each other to generate long filaments that are maintained by bound cOA ligands, with stacked TIR domains forming the NAD + cleavage catalytic sites. Further, Cat1 filaments assemble into unique trigonal and pentagonal networks that enhance NAD + degradation. Cat1 presents an unprecedented chemistry and higher-order protein assembly for the CRISPR-Cas response.
- Laboratory of Bacteriology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: