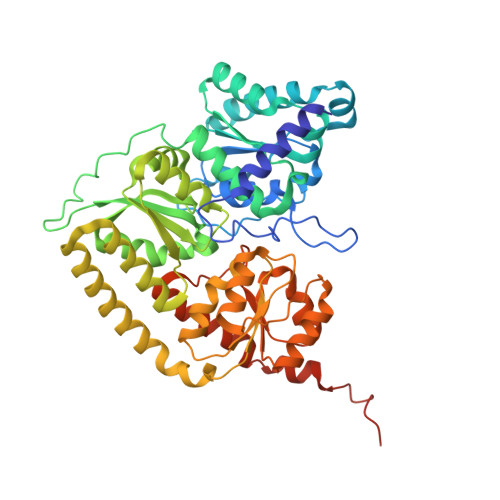
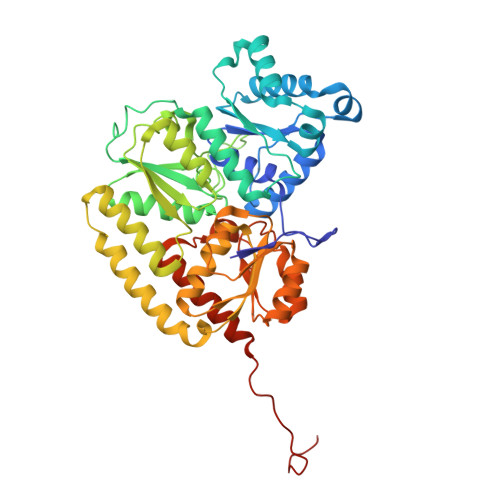
Dynamics driving the precursor in NifEN scaffold during nitrogenase FeMo-cofactor assembly.
Paya Tormo, L., Nguyen, T.Q., Fyfe, C., Basbous, H., Dobrzynska, K., Echavarri-Erasun, C., Martin, L., Caserta, G., Legrand, P., Thorn, A., Amara, P., Schoehn, G., Cherrier, M.V., Rubio, L.M., Nicolet, Y.(2025) Nat Chem Biol
- PubMed: 41238839
- DOI: https://doi.org/10.1038/s41589-025-02070-4
- Primary Citation of Related Structures:
9I0F, 9I0G, 9I0H - PubMed Abstract:
Nitrogenase catalyzes atmospheric nitrogen fixation, a critical biological process that depends on an intricate organometallic cofactor assembled by a dedicated multiprotein system. Here we uncover the structural basis for the function of NifEN, the scaffold protein that mediates the final stages of cofactor biosynthesis before its incorporation into nitrogenase. High-resolution structural analyses reveal that the cofactor precursor initially binds at a surface docking site before being transferred into a specialized cavity for further maturation. This process involves dynamic structural rearrangements, including coordinated domain motions and partial unfolding, enabling the scaffold to alternate between open and closed states. Additionally, a rear channel extends to the precursor-binding cavity, likely facilitating the entry of the modifying components molybdenum and homocitrate. These findings illuminate the dynamic mechanisms underlying FeMo-cofactor assembly and underscore the functional divergence between NifEN, the biosynthetic scaffold, and NifDK, the catalytic component of nitrogenase.
- Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid e Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria/Consejo Superior de Investigaciones Científicas, Madrid, Spain.
Organizational Affiliation: