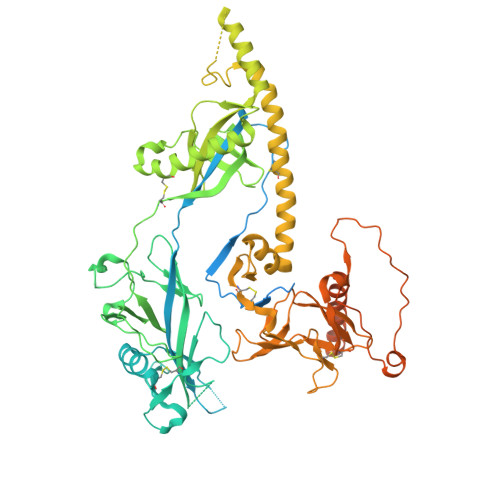
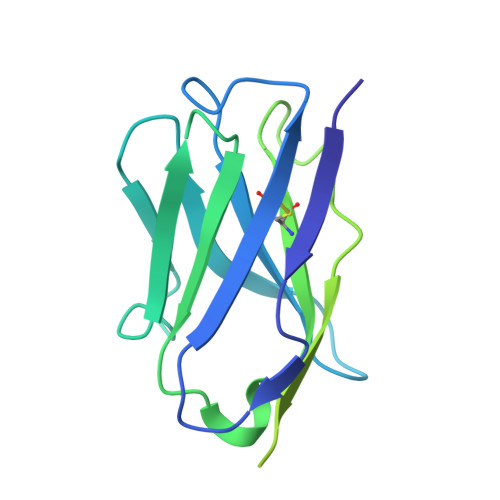
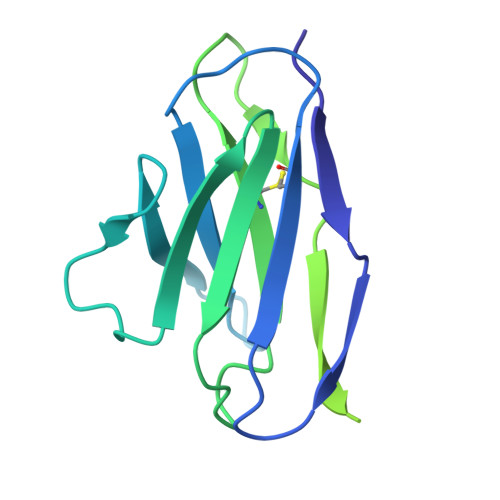
Prefusion structure, evasion and neutralization of HSV-1 glycoprotein B.
Roark, R.S., Schaub, A.J., Shi, W., Wang, M., Bahna, F.A., Becker, J.E., Biju, A., Chong, S., Du, H., Guo, Y., Hong, H., Katsamba, P.S., Mannepalli, S.M., Olia, A.S., Ou, L., Rubin, S.K., Sabo, Y., Suleiman, M., Wells, M.L., Zhang, B., Cheng, C., Glasgow, A., Ho, D.D., Huang, Y., Pierson, T.C., Rawi, R., Zhou, T., Shapiro, L., Kwong, P.D.(2025) Nat Microbiol 10: 2966-2980
- PubMed: 41174178
- DOI: https://doi.org/10.1038/s41564-025-02153-x
- Primary Citation of Related Structures:
9DD6, 9DD7, 9DD8, 9DDA, 9DDC - PubMed Abstract:
Glycoprotein B (gB) refolds between prefusion and postfusion conformations to facilitate herpesvirus entry into host cells. However, the isolation of prefusion-specific neutralizing antibodies, effective against other viral entry machines, has been challenging. Here we describe stabilization of the prefusion gB ectodomain from herpes simplex virus 1 (HSV-1), determine ectodomain structures at 2.9- to 4.1-Å resolution using cryogenic electron microscopy (cryo-EM) and isolate a prefusion-specific gB-neutralizing antibody termed WS.HSV-1.24. Murine immunization with gB stabilized in the prefusion conformation induced high titres of antibodies binding to both prefusion and postfusion gB, but-most notably-without measurable serum neutralization. Accessibility analysis revealed iso-surface exposure, with accessible surfaces on prefusion HSV-1 gB also exposed on postfusion gB. Structural analysis suggested substantial plasticity, with regions that refolded between pre- and postfusion conformations relegated to domain interfaces with limited accessibility; indeed, WS.HSV-1.24 recognized a domain-interface refolding region to facilitate neutralization. We propose that prefusion HSV-1 gB evades neutralization by most antibodies through an iso-surface display that is coupled to structural plasticity.
- Aaron Diamond AIDS Research Center, Columbia University Vagelos College of Physicians and Surgeons, New York, NY, USA.
Organizational Affiliation: