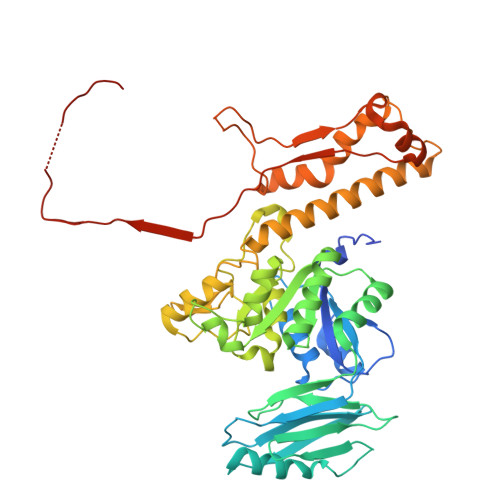
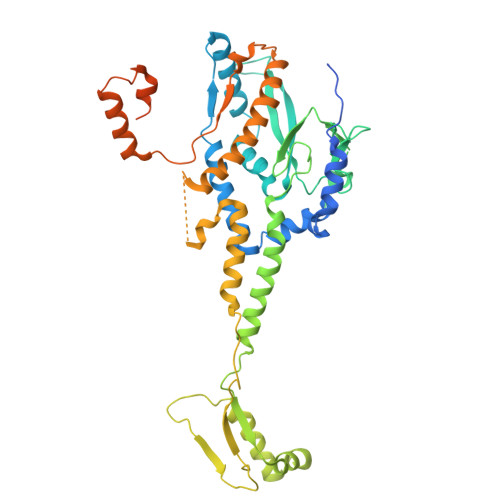
Global structural survey of the flagellotropic myophage phi TE infecting agricultural pathogen Pectobacterium atrosepticum.
Hodgkinson-Bean, J., Ayala, R., Jayawardena, N., Rutter, G.L., Watson, B.N.J., Mayo-Munoz, D., Keal, J., Fineran, P.C., Wolf, M., Bostina, M.(2025) Nat Commun 16: 3257-3257
- PubMed: 40188083
- DOI: https://doi.org/10.1038/s41467-025-58514-x
- Primary Citation of Related Structures:
9CB9, 9CBA, 9CC7, 9CUL, 9CUY, 9MJN - PubMed Abstract:
Bacteriophages offer a promising alternative to drug-based treatments due to their effectiveness and host specificity. This is particularly important in agriculture as a biocontrol agent of plant diseases. Phage engineering is facilitated by structural knowledge. However, structural information regarding bacteriophages infecting plant pathogens is limited. Here, we present the cryo-EM structure of bacteriophage φTE that infects plant pathogen Pectobacterium atrosepticum. The structure reveals a distinct neck topology compared with other myophages, where tail terminator proteins compensate for reduced connectivity between sheath subunits. A contact network between tail fibers, the sheath initiator, and baseplate wedge proteins provides insights into triggers that transduce conformational changes from the baseplate to the sheath to orchestrate contraction. We observe two distinct oligomeric states of the tape measure protein (TMP), which is six-fold in regions proximal to the N-terminus and throughout most of the tail, while three-fold at the C-terminus, indicating that the TMP may be proteolytically cleaved. Our results provide a structural atlas of the model bacteriophage φTE, enhancing future interpretation of phage host interactions in pectobacteria. We anticipate that our structure will inform rational design of biocontrol agents against plant pathogens that cause diseases such as soft rot and blackleg disease in potatoes.
- Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: