Structure and Mechanism of the Pyridoxal 5'-Phosphate-Dependent Amino Acid gamma-Substitution Enzyme in 6-Alkyl-Pipecolate Biosynthesis
Gao, Y., Wang, B., Zhou, J., Gu, Y.(2025) ACS Catal : 18893-18907
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PLP dependent gamma-lyase CndF | A [auth B], B [auth A] | 568 | Penicillium citrinum | Mutation(s): 0 | 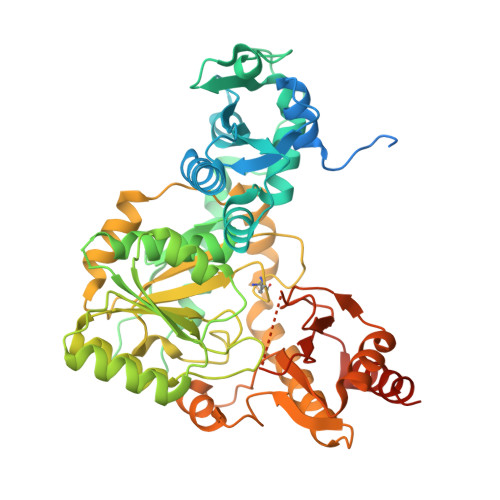 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 Query on PO4 | C [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| CL Query on CL | D [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LLP Query on LLP | A [auth B], B [auth A] | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 85.432 | α = 90 |
| b = 102.527 | β = 90 |
| c = 128.124 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |