Crystal structure of xylitol-bound glucose isomerase by serial millisecond crystallography
Nam, K.H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Xylose isomerase | 388 | Streptomyces pseudogriseolus | Mutation(s): 0 EC: 5.3.1.5 | 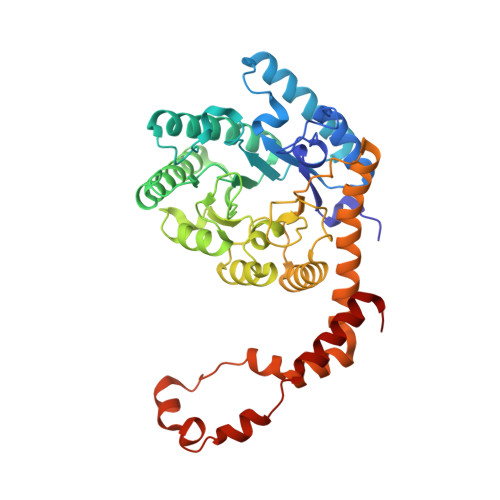 | |
UniProt | |||||
Find proteins for P24300 (Streptomyces rubiginosus) Explore P24300 Go to UniProtKB: P24300 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P24300 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XYL (Subject of Investigation/LOI) Query on XYL | C [auth A] | Xylitol C5 H12 O5 HEBKCHPVOIAQTA-SCDXWVJYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | B [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 94.138 | α = 90 |
| b = 99.936 | β = 90 |
| c = 103.156 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| CrystFEL | data reduction |
| CrystFEL | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Korea) | Korea, Republic Of | NRF-2017M3A9F6029736 |
| National Research Foundation (NRF, Korea) | Korea, Republic Of | NRF-2017R1D1A1B03033087 |